In this post I will explain how to sync a PostgreSQL table to an Elasticsearch index using Debezium. This is a follow-up on my previous post. In that post I explained how to load a GBIF dataset into a PostgreSQL database.
Debezium’s PostgreSQL connector captures row-level changes in the schemas of a PostgreSQL database. This technique is called Change Data Capture (CDC) and can be used to sync a database table to (in this case) Elasticsearch. CDC makes it possible to move heavy search workloads away from the database.
In this example I’m going to explain how to sync a table named explore.gbif to an Elasticsearch index named db1.explore.gbif, using the following components:

Download this docker-compose project to try this setup on your local computer using Docker. After starting the containers the dataset will be downloaded and inserted into a PostgreSQL table.
When all containers are up, run the sync_to_elasticsearch.sh script. This script will do the following:
- Create an index and field mappings
- Setup the PostgreSQL source connector
- Setup the Elasticsearch sink connector
There are three fields that need a mapping, gbifid, eventdate and location. The location field is used to plot locations on a world map in Kibana:
{
"settings": {
"number_of_shards": 1
},
"mappings": {
"properties": {
"gbifid": {
"type": "long"
},
"eventdate": {
"type": "date"
},
"location": {
"type": "geo_point"
}
}
}
}
The psql connector is used to create the connection between kafka and the database. Notice the column.blacklist setting to exclude the PostGIS geometry and tsv columns.
{
"name": "psql_connector",
"config": {
"connector.class": "io.debezium.connector.postgresql.PostgresConnector",
"tasks.max": "1",
"database.hostname": "postgres",
"database.port": "5432",
"database.user": "postgres",
"database.password": "postgres",
"database.dbname": "db1",
"database.server.name": "db1",
"database.whitelist": "db1",
"heartbeat.interval.ms": "1000",
"table.whitelist": "explore.gbif",
"column.blacklist":"explore.gbif\\.(geom|tsv).*",
"database.history.kafka.bootstrap.servers": "kafka:9092",
"plugin.name": "pgoutput"
}
}
The Elasticsearch connector is used to connect kafka to the Elasticsearch instance. The transforms.key.field is set to gbifid, the primary key in the table.
{
"name": "es_connector",
"config": {
"connector.class": "io.confluent.connect.elasticsearch.ElasticsearchSinkConnector",
"tasks.max": "1",
"topics": "db1.explore.gbif",
"connection.url": "http://elasticsearch:9200",
"transforms": "unwrap,key",
"transforms.unwrap.type": "io.debezium.transforms.UnwrapFromEnvelope",
"transforms.unwrap.drop.tombstones": "false",
"transforms.unwrap.drop.deletes": "false",
"transforms.key.type": "org.apache.kafka.connect.transforms.ExtractField$Key",
"transforms.key.field": "gbifid",
"key.ignore": "false",
"type.name": "_doc",
"behavior.on.null.values": "delete"
}
}
Using the docker-compose logs command to see that the table is being synced to the Elasticsearch index:
Use the following command to see when the sync is finished:
docker-compose logs -f connect
docker-compose logs connect | grep 'Finished exporting'
A log line should like this should show after a couple of minutes:
Finished exporting 287671 records for table 'explore.gbif'; total duration '00:01:41.021' [io.debezium.relational.RelationalSnapshotChangeEventSource]
Now let’s check if all records are synced to Elasticsearch. Query the number of records in the Elasticsearch index:
curl http://localhost:9200/db1.explore.gbif/_count
{
"count": 287671,
"_shards": {
"total": 1,
"successful": 1,
"skipped": 0,
"failed": 0
}
}
Query the number of records in the table. Both queries show the same number, this confirms that all records are correctly synced:
docker-compose exec postgres bash -c 'psql -U $POSTGRES_USER $POSTGRES_DB -c "SELECT COUNT(*) FROM explore.gbif"' count -------- 287671 (1 row)
The next step is to load a Kibana dashboard using the script called load_kibana_dashboard.sh. Run this script. The output of this script should look like this:
Load Kibana dashboard
{"success":true,"successCount":7}
Now browse to http://localhost:5601/app/kibana#/dashboards. Click on the dashboard called Explore a GBIF dataset.
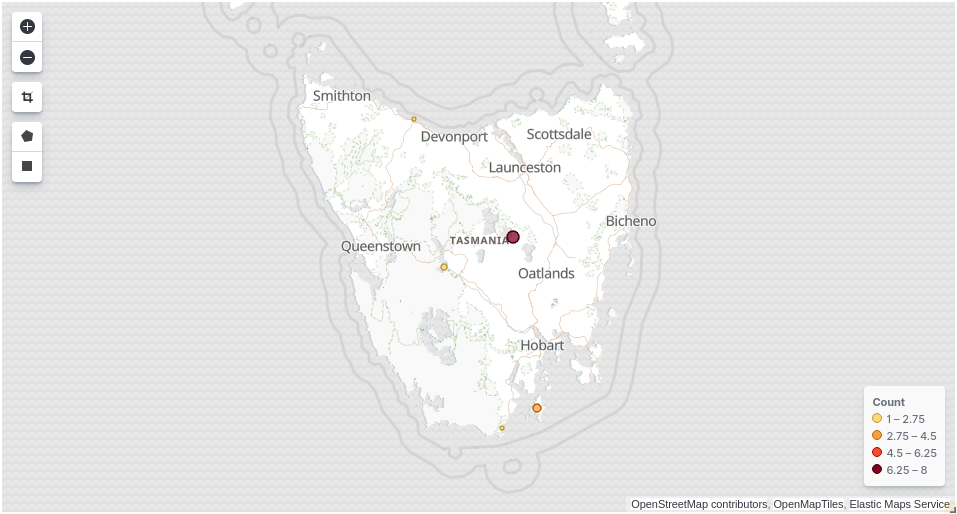
Compare the map in Kibana (above) with the map in Grafana (below). As you can see the data is identical.

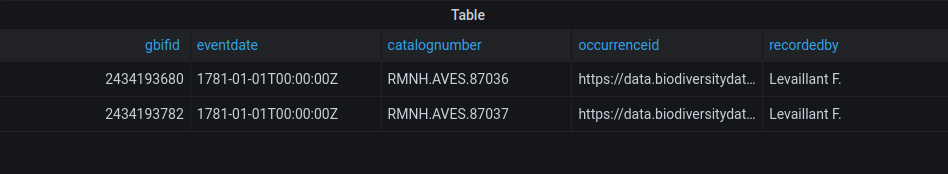
Record with gbifid 2434193680 in the database:
SELECT * FROM explore.gbif WHERE gbifid IN (2434193680): gbifid | 2434193680 eventdate | 1781-01-01 00:00:00+00 year | 1781 month | 1 day | 1 catalognumber | RMNH.AVES.87036 occurrenceid | https://data.biodiversitydata.nl/naturalis/specimen/RMNH.AVES.87036 recordedby | Levaillant F. sex | FEMALE lifestage | JUVENILE preparations | mounted skin locality | stateprovince | Cape of Good Hope countrycode | ZA higherclassification | Phalacrocoracidae kingdom | Animalia phylum | Chordata class | Aves order | Suliformes family | Anhingidae genus | Anhinga specificepithet | melanogaster species | Anhinga rufa genericname | Anhinga scientificname | Anhinga melanogaster rufa (Daudin, 1802) decimallatitude | -34.0083 decimallongitude | 19.0083 geom | 0101000020E61000008A8EE4F21F023340454772F90F0141C0 location | -34.0083,19.0083
The same record in the Elasticsearch index:
GET /db1.explore.gbif/_search?q=gbifid:2434193680
{
"took" : 22,
"timed_out" : false,
"_shards" : {
"total" : 1,
"successful" : 1,
"skipped" : 0,
"failed" : 0
},
"hits" : {
"total" : {
"value" : 1,
"relation" : "eq"
},
"max_score" : 1.0,
"hits" : [
{
"_index" : "db1.explore.gbif",
"_type" : "_doc",
"_id" : "2434193680",
"_score" : 1.0,
"_source" : {
"gbifid" : 2434193680,
"eventdate" : "1781-01-01T00:00:00Z",
"year" : 1781,
"month" : 1,
"day" : 1,
"catalognumber" : "RMNH.AVES.87036",
"occurrenceid" : "https://data.biodiversitydata.nl/naturalis/specimen/RMNH.AVES.87036",
"recordedby" : "Levaillant F.",
"sex" : "FEMALE",
"lifestage" : "JUVENILE",
"preparations" : "mounted skin",
"locality" : null,
"stateprovince" : "Cape of Good Hope",
"countrycode" : "ZA",
"higherclassification" : "Phalacrocoracidae",
"kingdom" : "Animalia",
"phylum" : "Chordata",
"class" : "Aves",
"order" : "Suliformes",
"family" : "Anhingidae",
"genus" : "Anhinga",
"specificepithet" : "melanogaster",
"species" : "Anhinga rufa",
"genericname" : "Anhinga",
"scientificname" : "Anhinga melanogaster rufa (Daudin, 1802)",
"decimallatitude" : -34.0083,
"decimallongitude" : 19.0083,
"location" : "-34.0083,19.0083"
}
}
]
}
}
Happy exploring!